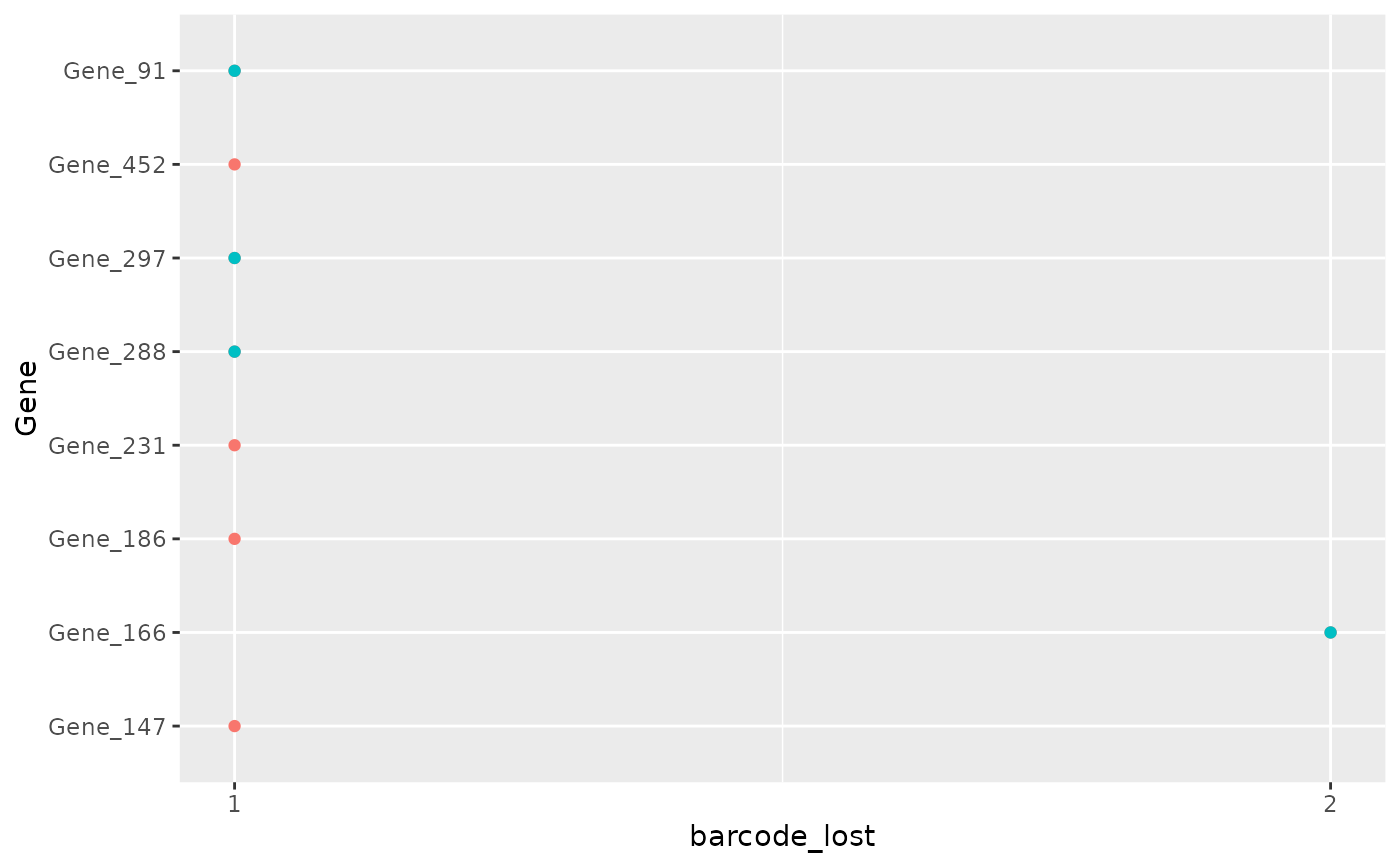
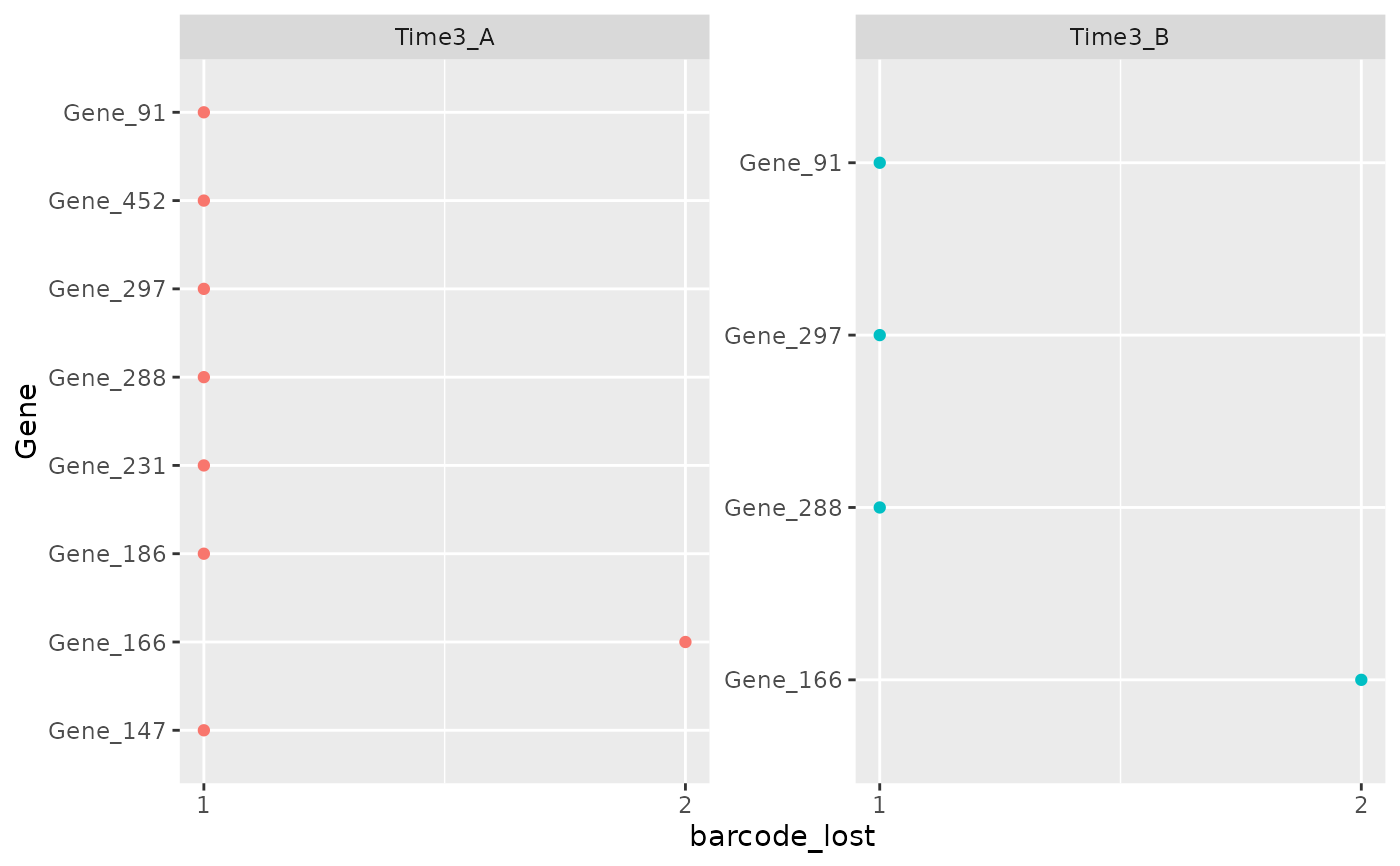
This function plots the number of barcodes lost in each sample for each gene. Usually in a genetic screening each gene is is associated with multiple shRNAs and so barcodes. For this reason a reasonable number of barcodes associated with the gene has to be retrieved in order to have a robust result. Visualizing the number of genes that have lost lot's of barcode is a Quality Check procedure in order to be aware of the number of barcode for the hit identified.
Arguments
- screenR_Object
The ScreenR object obtained using the
create_screenr_object- facet
A boolean to use the facet.
- samples
A vector of samples that as to be visualize
Examples
object <- get0("object", envir = asNamespace("ScreenR"))
plot_barcode_lost_for_gene(object,
samples = c("Time3_A", "Time3_B")
)
 plot_barcode_lost_for_gene(object,
samples = c("Time3_A", "Time3_B"),
facet = FALSE
)
plot_barcode_lost_for_gene(object,
samples = c("Time3_A", "Time3_B"),
facet = FALSE
)